-Search query
-Search result
Showing 1 - 50 of 86 items for (author: cronin & n)

EMDB-19079: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 5nt pre-translocated complex
Method: single particle / : Gao F, Zhang X

EMDB-19080: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 5nt post-translocated complex
Method: single particle / : Gao F, Zhang X

EMDB-19081: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 6nt complex
Method: single particle / : Gao F, Zhang X

EMDB-19082: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 7nt complex
Method: single particle / : Gao F, Zhang X

EMDB-19083: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 8nt complex
Method: single particle / : Gao F, Zhang X

EMDB-19084: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 9nt complex
Method: single particle / : Gao F, Zhang X

PDB-8re4: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 5nt pre-translocated complex
Method: single particle / : Gao F, Zhang X

PDB-8rea: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 5nt post-translocated complex
Method: single particle / : Gao F, Zhang X

PDB-8reb: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 6nt complex
Method: single particle / : Gao F, Zhang X

PDB-8rec: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 7nt complex
Method: single particle / : Gao F, Zhang X

PDB-8red: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 8nt complex
Method: single particle / : Gao F, Zhang X

PDB-8ree: 
Cryo-EM structure of bacterial RNA polymerase-sigma54 initial transcribing complex - 9nt complex
Method: single particle / : Gao F, Zhang X

EMDB-17169: 
Cryo-EM structure of the SARS-CoV-2 Spike bound to TMEM106B
Method: single particle / : CHEREPANOV P

EMDB-17170: 
Local refinement of the SARS-CoV-2 Spike RBD bound to TMEM106B
Method: single particle / : CHEREPANOV P

EMDB-16914: 
Cryo-EM structure of the DnaD-NTD tetramer
Method: single particle / : Winterhalter C, Pelliciari S, Cronin N, Costa TRD, Murray H, Ilangovan A

PDB-8ojj: 
Cryo-EM structure of the DnaD-NTD tetramer
Method: single particle / : Winterhalter C, Pelliciari S, Cronin N, Costa TRD, Murray H, Ilangovan A

EMDB-13663: 
CryoEM structure of DnaD dimer from Bacillus subtilis
Method: single particle / : Winterhalter C, Pelliciari S, Cronin N, Costa TRD, Ilangovan I, Murray H

EMDB-13895: 
CryoEM structure of the Smc5/6-holocomplex (composite structure)
Method: single particle / : Hallett ST, Oliver AW

PDB-7qcd: 
CryoEM structure of the Smc5/6-holocomplex (composite structure)
Method: single particle / : Hallett ST, Oliver AW

EMDB-13893: 
Cryo-EM structure of the Smc5/6 holo-complex; map for head-end of complex.
Method: single particle / : OLIVER AW, Hallett ST

EMDB-13894: 
CryoEM structure of the Smc5/6 holocomplex; map for hinge and arm region.
Method: single particle / : OLIVER AW, Hallett ST

EMDB-14591: 
CryoEM structure of SARS-CoV-2 spike monomer in complex with neutralising antibody P008_60
Method: single particle / : Rosa A, Pye VE, Cronin N, Cherepanov P

PDB-7zbu: 
CryoEM structure of SARS-CoV-2 spike monomer in complex with neutralising antibody P008_60
Method: single particle / : Rosa A, Pye VE, Cronin N, Cherepanov P
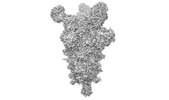
EMDB-26676: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26677: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-26678: 
An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-27043: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody SP1-77 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K
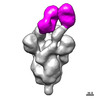
EMDB-27044: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-7-53 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

EMDB-27046: 
Negative stain electron microscopy single particle reconstruction of monoclonal antibody VHH7-5-82 Fab in complex with SARS-CoV2 2P spike
Method: single particle / : Edwards RJ, Mansouri K

PDB-7upw: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

PDB-7upx: 
Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains)
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

PDB-7upy: 
An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion
Method: single particle / : Zhang J, Luo S, Kreutzberger A, Kirchhausen T, Chen B, Haynes B, Alt F

EMDB-25105: 
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Gobeil S, Acharya P

PDB-7sg4: 
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Gobeil S, Acharya P

EMDB-13075: 
Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450
Method: single particle / : Barski MS, Ballandras-Colas A, Cronin NB, Pye VE, Cherepanov P, Maertens GN

EMDB-13076: 
STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir
Method: single particle / : Barski MS, Ballandras-Colas A, Cronin NB, Pye VE, Cherepanov P, Maertens GN

EMDB-13077: 
Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir
Method: single particle / : Barski MS, Ballandras-Colas A, Cronin NB, Pye VE, Cherepanov P, Maertens GN

PDB-7ouf: 
Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450
Method: single particle / : Barski MS, Ballandras-Colas A, Cronin NB, Pye VE, Cherepanov P, Maertens GN

PDB-7oug: 
STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir
Method: single particle / : Barski MS, Ballandras-Colas A, Cronin NB, Pye VE, Cherepanov P, Maertens GN

PDB-7ouh: 
Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir
Method: single particle / : Barski MS, Ballandras-Colas A, Cronin NB, Pye VE, Cherepanov P, Maertens GN

EMDB-23400: 
SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab
Method: single particle / : Gobeil S, Acharya P

PDB-7ljr: 
SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab
Method: single particle / : Gobeil S, Acharya P

EMDB-23246: 
CryoEM map of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1041
Method: single particle / : Manne K, Acharya P

PDB-7laa: 
Structure of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1041
Method: single particle / : Manne K, Acharya P

EMDB-23248: 
CryoEM map of SARS-CoV-2 S protein in complex with N-terminal domain antibody DH1052
Method: single particle / : Manne K, Acharya P

PDB-7lab: 
Structure of SARS-CoV-2 S protein in complex with N-terminal domain antibody DH1052
Method: single particle / : Manne K, Acharya P

EMDB-23277: 
CryoEM map of SARS-CoV-2 S protein in complex with N-terminal domain antibody DH1050.1
Method: single particle / : Manne K, Acharya P

EMDB-23279: 
CryoEM map of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Manne K, Acharya P

PDB-7lcn: 
Structure of SARS-CoV-2 S protein in complex with N-terminal domain antibody DH1050.1
Method: single particle / : Manne K, Acharya P

PDB-7ld1: 
Structure of SARS-CoV-2 S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Manne K, Acharya P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
